Getting started with Digest
Digest allows users to design MRM assays in an end-to-end manner, using its digestion, peptide query, and MRM design modules.
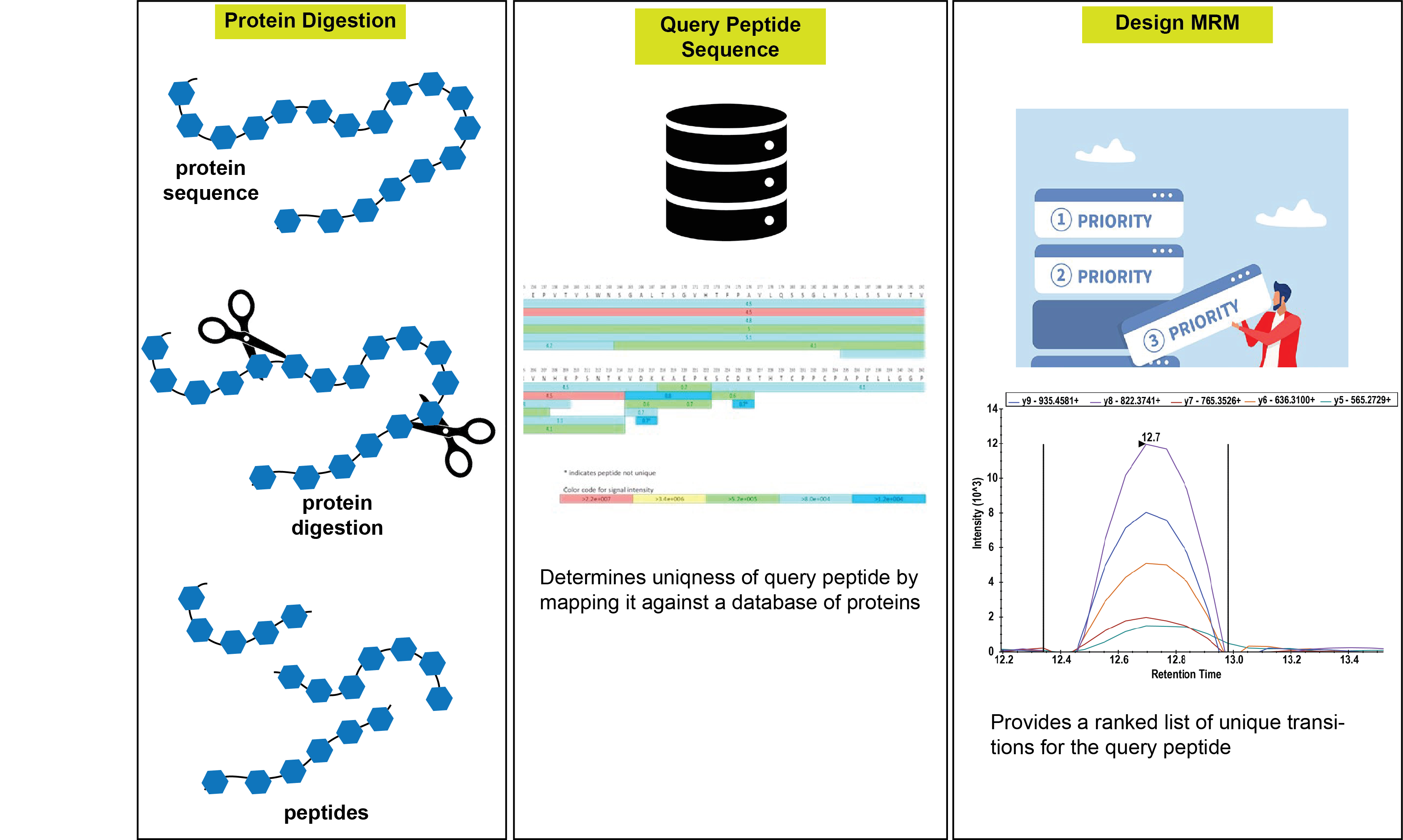
The typical workflow for the same is as follows:
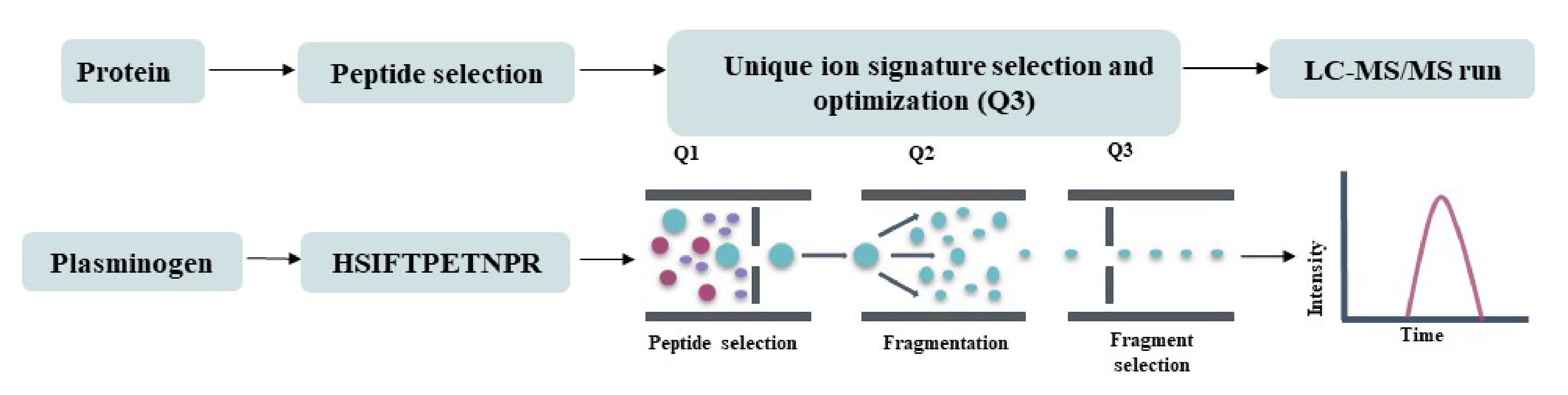
1. The User identifies the protein of their interest and obtains the query protein sequence from Uniprot. In this example, we choose human plasminogen (Uniprot ID: P00747).
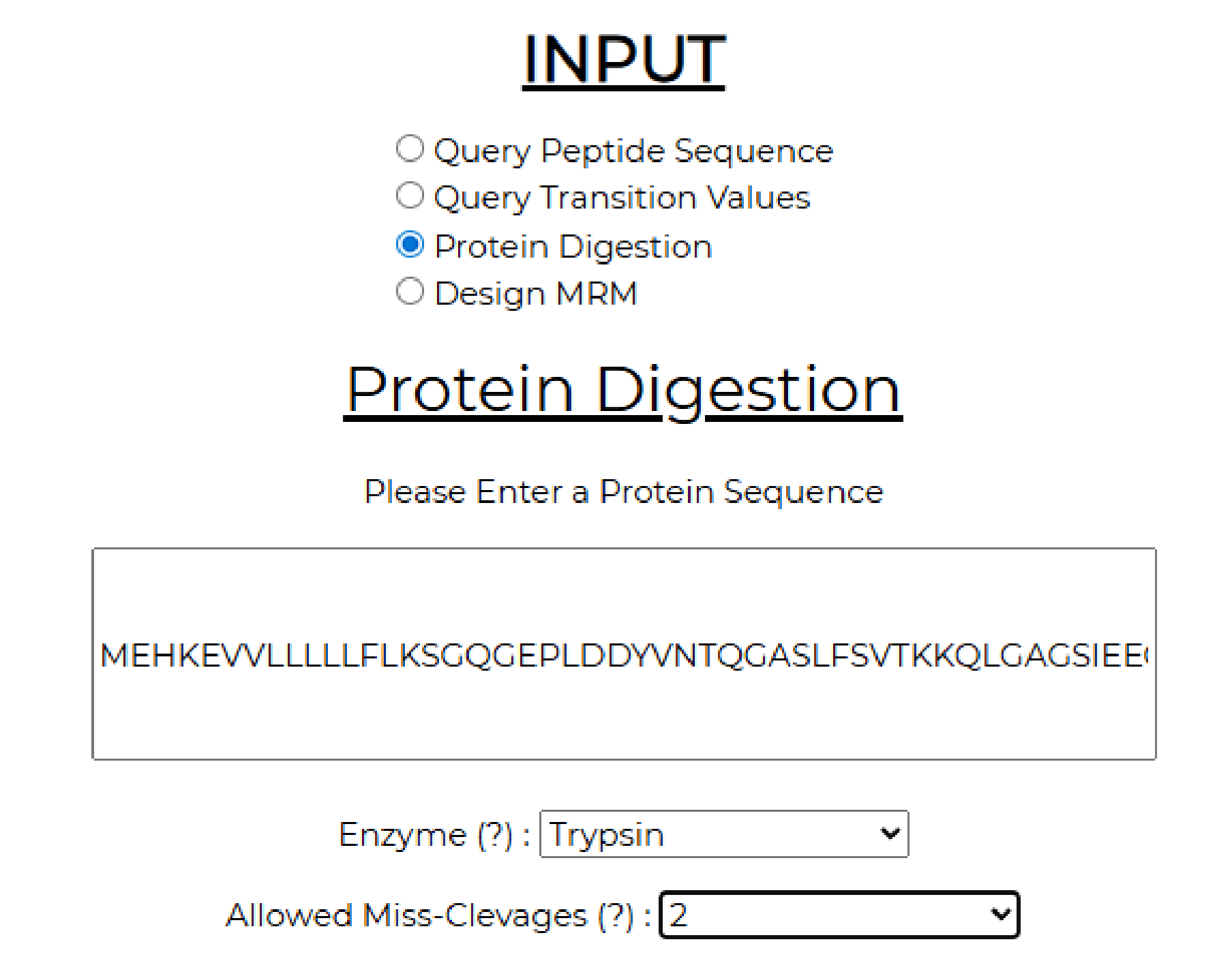
2. The input sequence is entered into the protein digestion module. The user chooses the required number of miscleavages and the enzyme for digestion. In this example, we choose up to 2 miscleavages and trypsin.
3. Digest provides an output table containing all the theoretical peptide fragments, along with their corresponding masses, and the number of miscleavages.
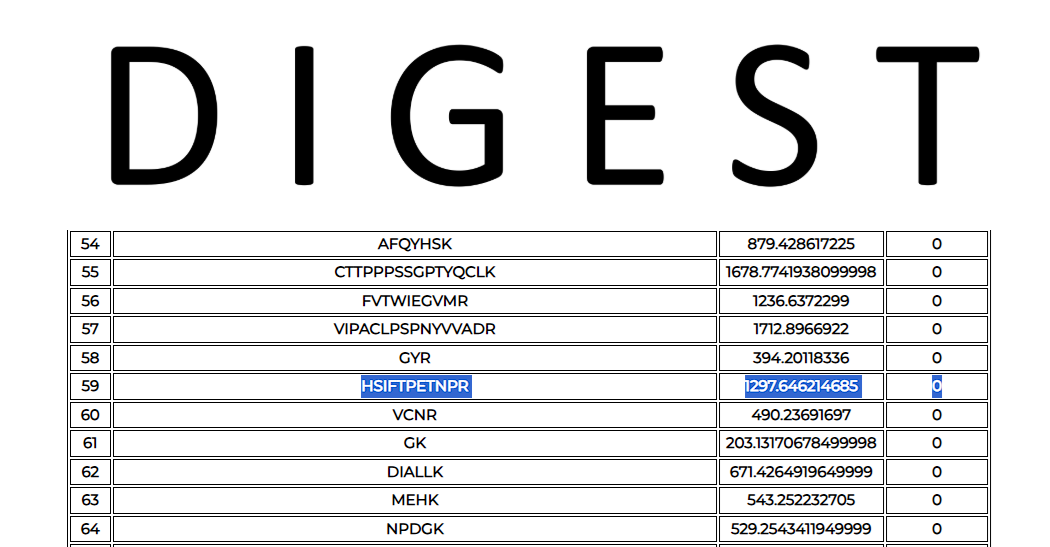
From this list, the user can pick a peptide against which they wish to design an MRM assay. The choice may depend on the probability of generating the peptide (as determined by the ExPASy peptide cutter) and its uniqueness.
4. Digest can be used to verify the uniqueness of the chosen peptide by determining how many proteins within an organism (or across organisms) it maps to through its query peptide sequence module.
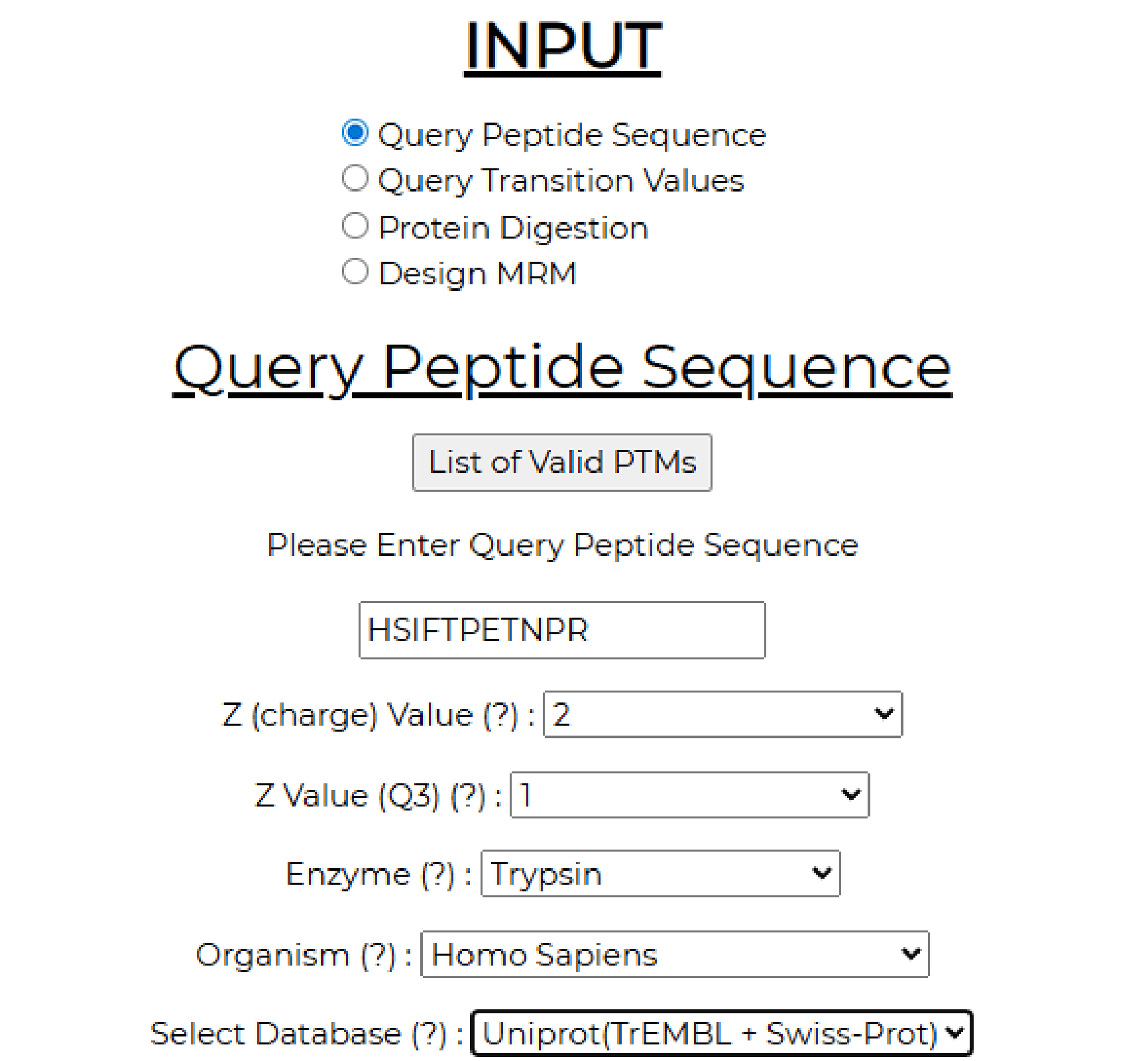
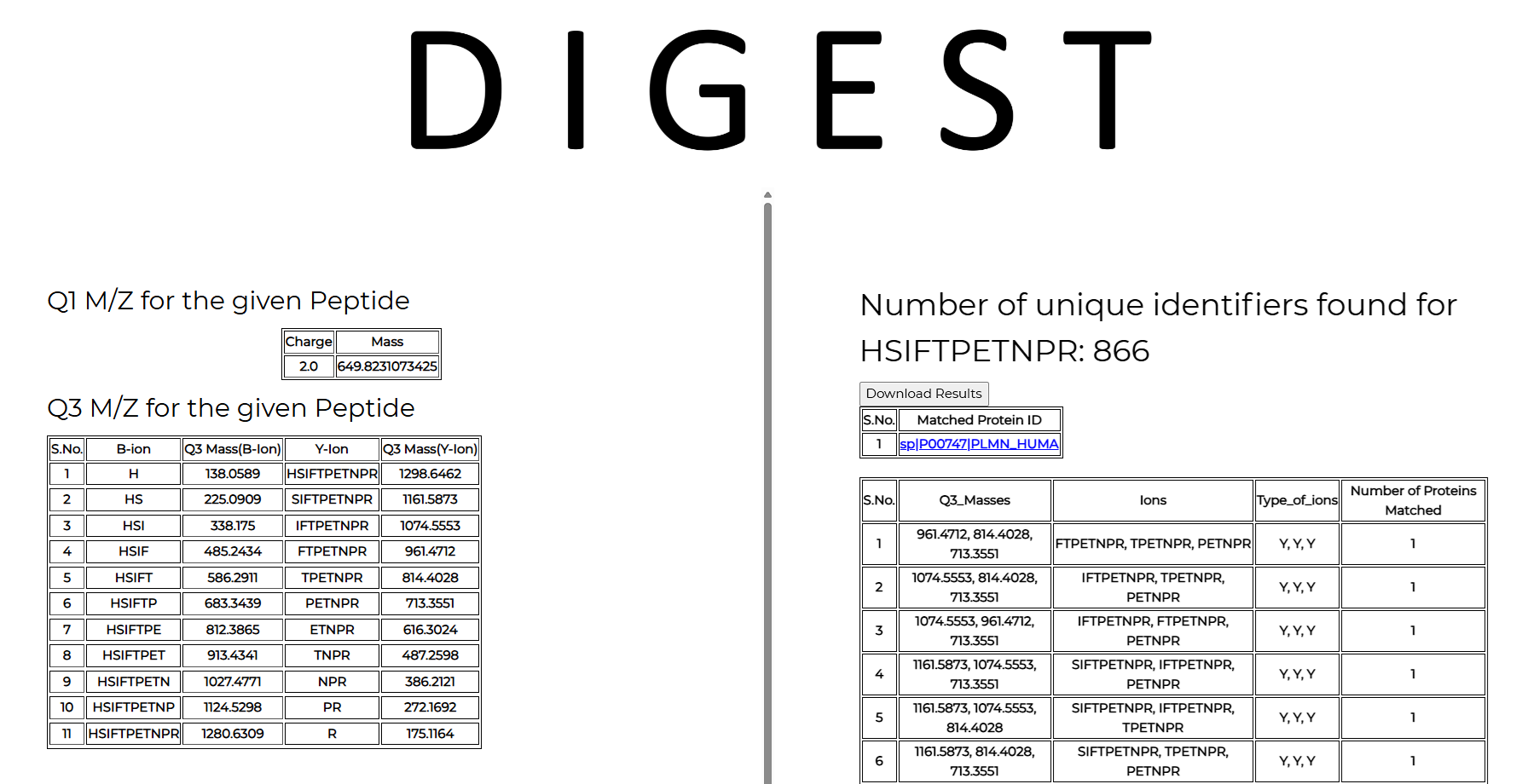
5. Query peptide sequence requires the peptide sequence input (in this example: HSIFTPETNPR) with q1 and q3 charges set to 2 and 1, respectively. The enzyme used for digestion (Trypsin), the organism, and the database to be used for protein mapping (Uniprot for this example).
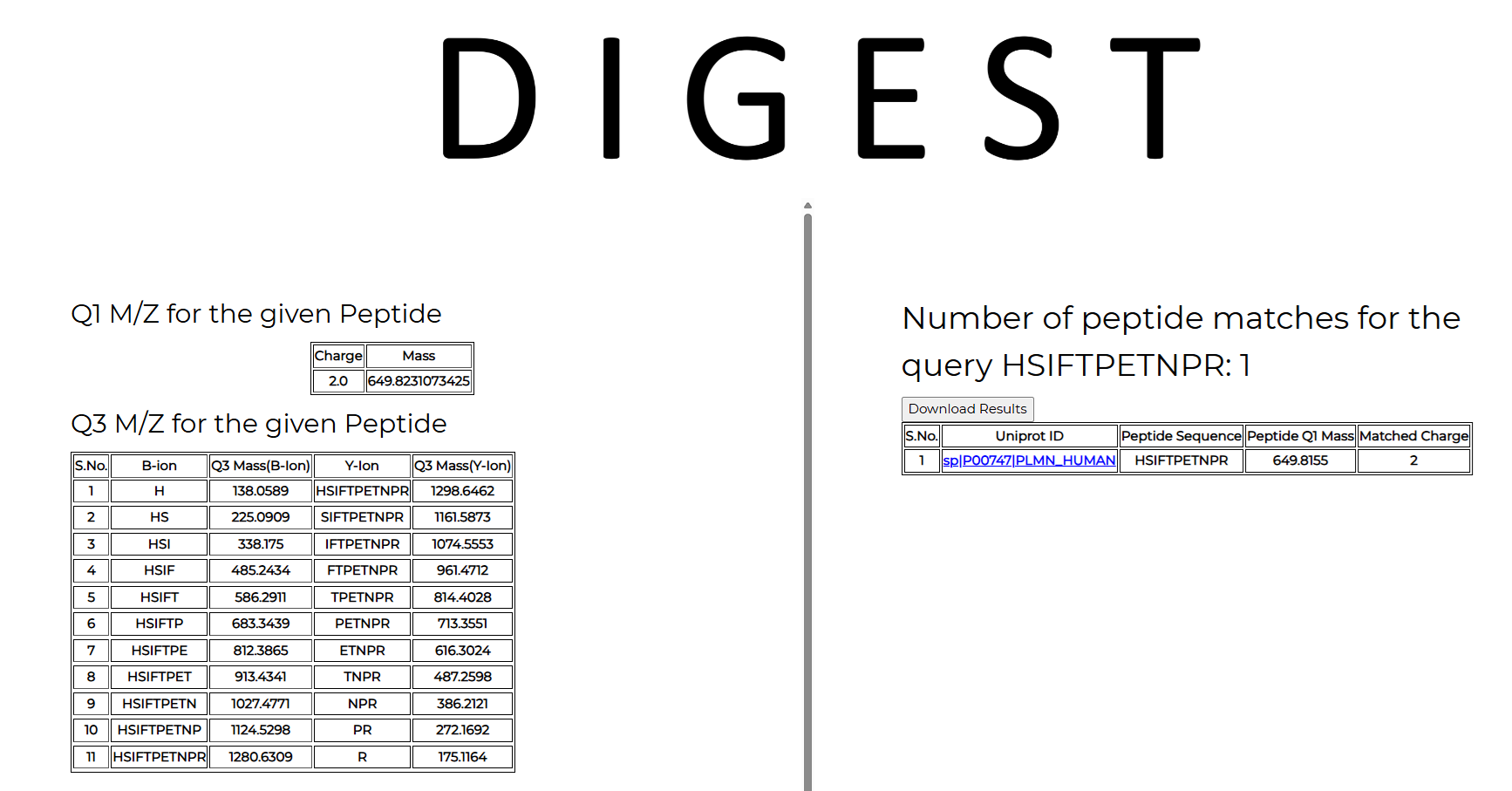
The output lists two tables. On the left of the output page is a table that contains all the Q3 ions generated from the given Q1 query peptide, along with their type (b or y-ion) and Q3 masses. On the right side of the output page is a table containing all the matches obtained from the chosen database for the given query.
In the current example, the chosen peptide HSIFTPETNPR maps to only one protein in the UniProt+Tremble database, with a Q1 mass of 649.8155 and a charge of 2. The link directs you to the UniProt website, where you can obtain additional details about the protein. The peptide is unique and only maps to human plasminogen.
6. To obtain transitions for this peptide that can be used in an MRM assay, the Design MRM module is utilized, requiring the peptide sequence (HSIFTPETNPR), Q1 and Q3 charges (2 and 1, respectively), instrument tolerance (0.5), enzyme used for digestion (trypsin), organism (Homo sapiens), and database for search (Uniprot) as inputs.
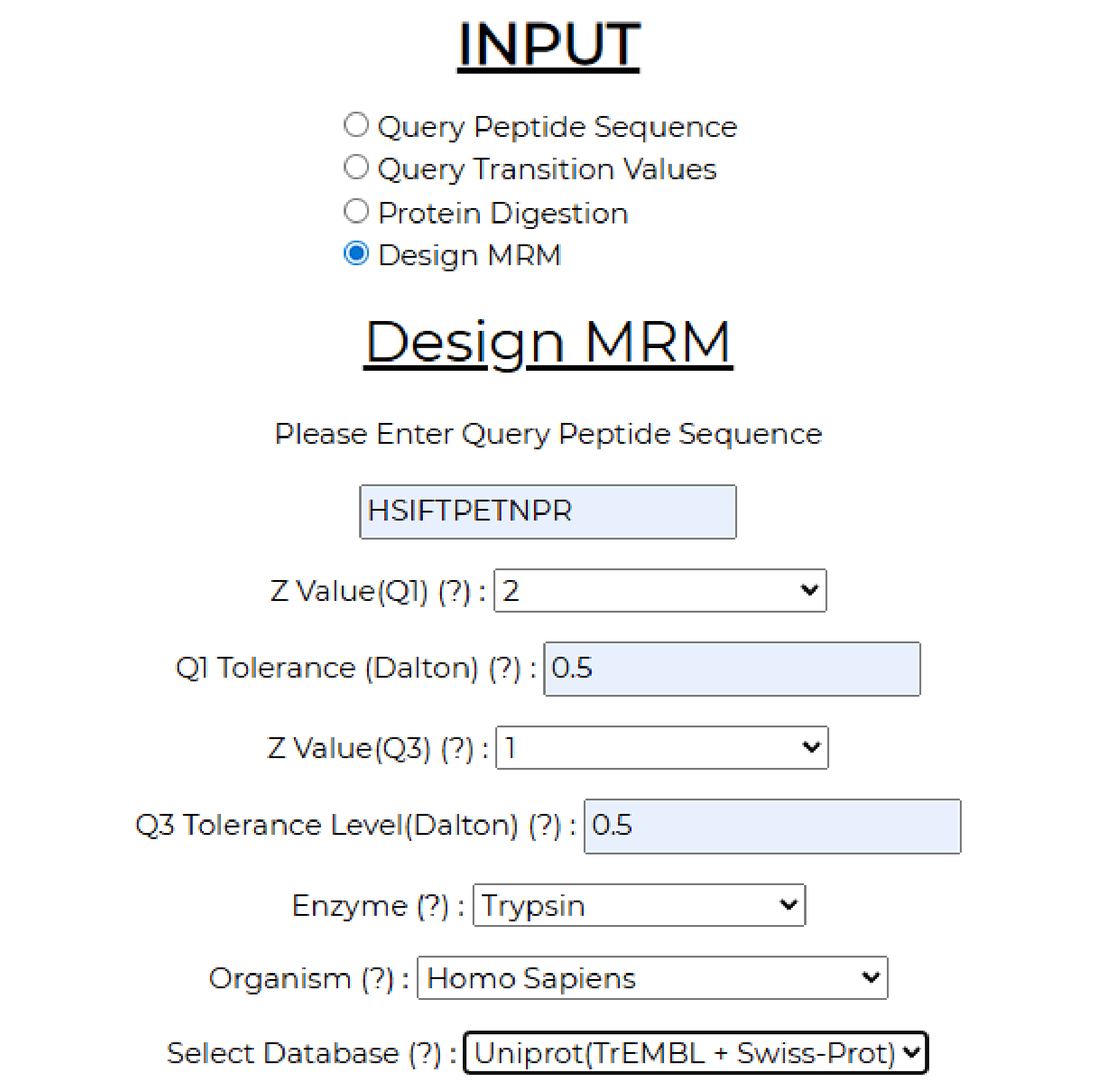
The output page displays two tables in a similar format. On the left is a table that contains all the Q3 ions generated from the given Q1 query peptide, along with their type (b or y-ion) and Q3 masses. On the right is a table listing combinations of three transitions (Q3 ions) that uniquely map to this peptide. These combinations can be directly used in MRM methods to quantify the abundance of the target protein.